| FusionGDB | FusionGDB2 | FusionPDB | FusionNeoAntigen | FGviewer | FusionAI | FusionNW |
FGviewer is aiming for the effective visualization of the functional features of fusion genes for better
prioritization of therapeutic target selection. FGviewer provides functional feature annotations at four different levels: DNA-, RNA-,
protein-, and pathogenic-levels. The same breakpoint line across four tiers will classify between FG involving
or non-involving zone with multiple types of functional features. Those features include fusion mRNA and
amino acid sequences based on the user’s breakpoint coordinates, swapped gene expression regulatory
(i.e., transcription factor or miRNA binding sites), protein functional features (i.e., protein domains, protein-
protein interactions, binding sites of all molecules, secondary structure level feature, etc.), clinically relevant
variants, etc.
*** Update: we corrected the error relating to the human genome version confusion (March 17, 2021).

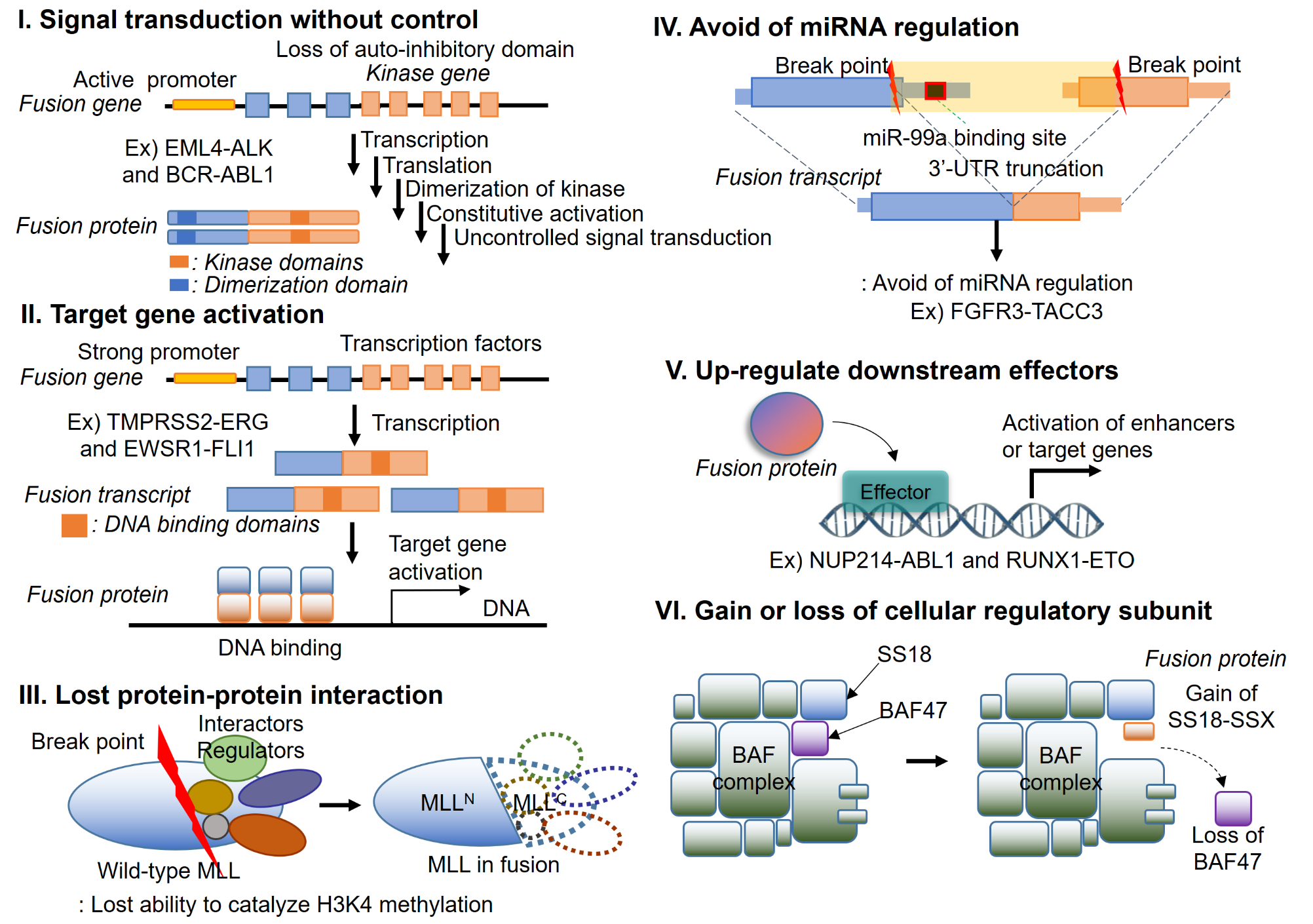
Understanding the function of fusion genes for tumorigenesis
Fusion genes can participate in gene regulation in a variety of ways in cancer cells. If a partner gene, with the active promoter and dimerization functional domains, is fused with a kinase domain, it will constitutively drive the activated expression of the kinase, leading to uncontrolled cell proliferation. For example, ALK is not normally transcribed in adult lung, but under the control of the EML4 promoter, the EML4-ALK fusion gene is transcribed. The dimerization domain of EML4 permits unregulated dimerization of the tyrosine kinase domain, constitutively activating downstream pathways. The BCR-ABL1 fusion gene is formed between BCR and the non-receptor tyrosine kinase, ABL1. Lacking auto-inhibitory N-terminal myristoylation in ABL1 and dimerization function of the coiled-coil domain in BCR contribute to the constitutive activation of kinase function. TMPRSS2-ERG fusion gene has over 50% of frequency in prostate cancer patients. The strong promoter of TMPRSS2, which is a prostate tissue-specific and androgen-inducing gene, enhanced the function of proto-oncogene (ERG). Similarly, if a DNA-binding domain of FLI1 is fused with a transactivation domain of EWSR1, this fusion protein can recruit transcriptional coregulatory elements, resulting in target gene activation. Through fusion, one protein can also lose its interactions with important cellular regulators (i.e., MLL fusion genes). If the 3’-UTR region of a fusion gene is truncated during fusion, such fusion transcript can avoid the regulation by its targeting miRNAs (i.e., FGFR3-TACC3). Other FG mechanisms include the loss of function of tumor suppressors and DNA damage repairs through the deletion or frameshift open reading frame (ORF) of fusion transcripts. Recently, several studies were done for identifying downstream effectors of FGs (i.e., NUP214-ABL1, RUNX1-ETO, and SS18-SSX). SS18, which encodes the BAF complex subunit, fused with SSX and affected the chromatin accessibility.